Software and Tools Developed by this Lab:
ConTra V3
A tool to identify transcription factor binding sites across species.
http://bioit2.irc.ugent.be/contra/v3
dynverse
dynverse is a collection of R packages aimed at supporting the trajectory inference (TI) community on multiple levels: end-users who want to apply TI on their dataset of interest, and developers who seek to easily quantify the performance of their TI method and compare it to other TI methods
https://dynverse.org/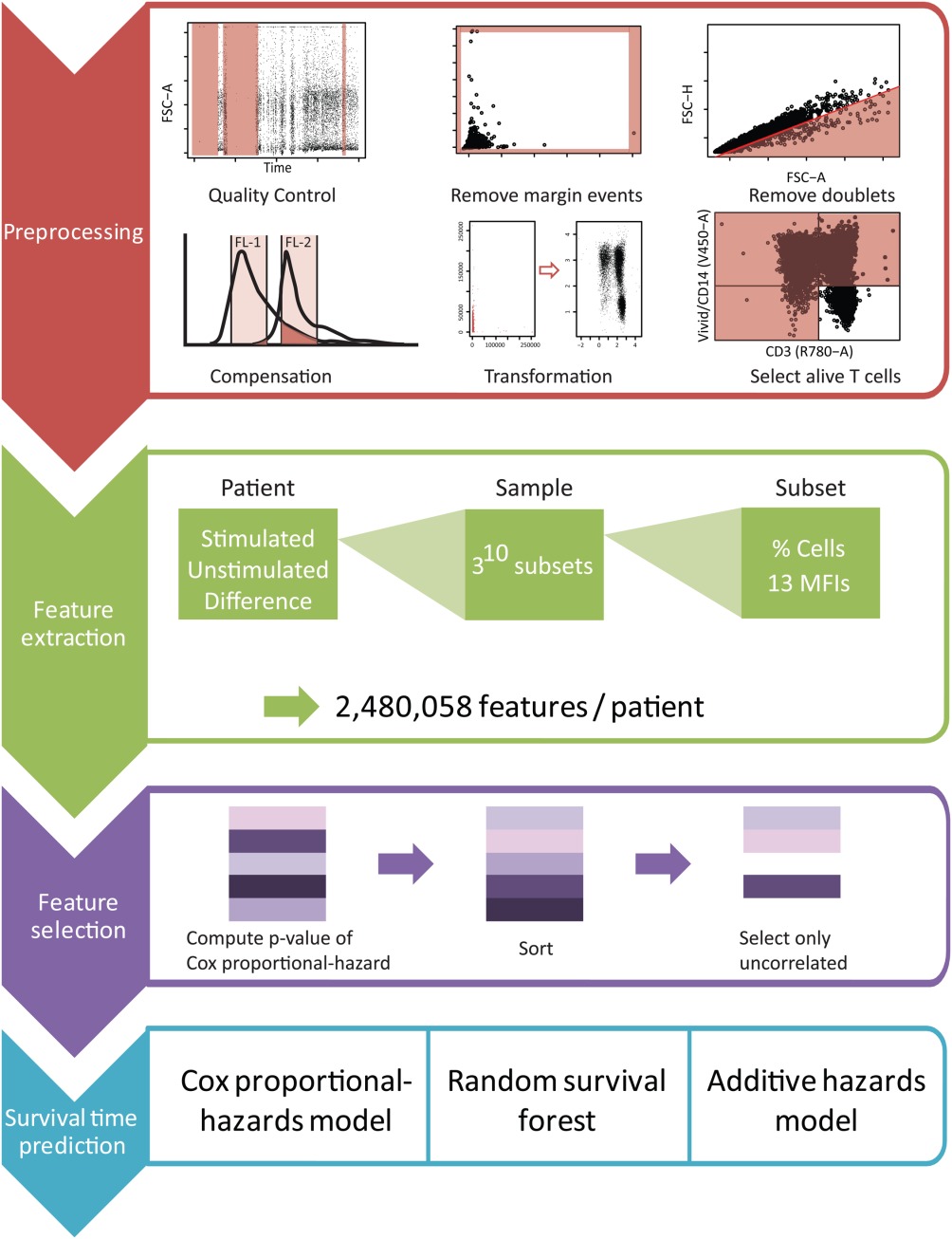
FloReMi
Flow density survival regression using minimal feature redundancy
https://github.com/saeyslab/FloReMi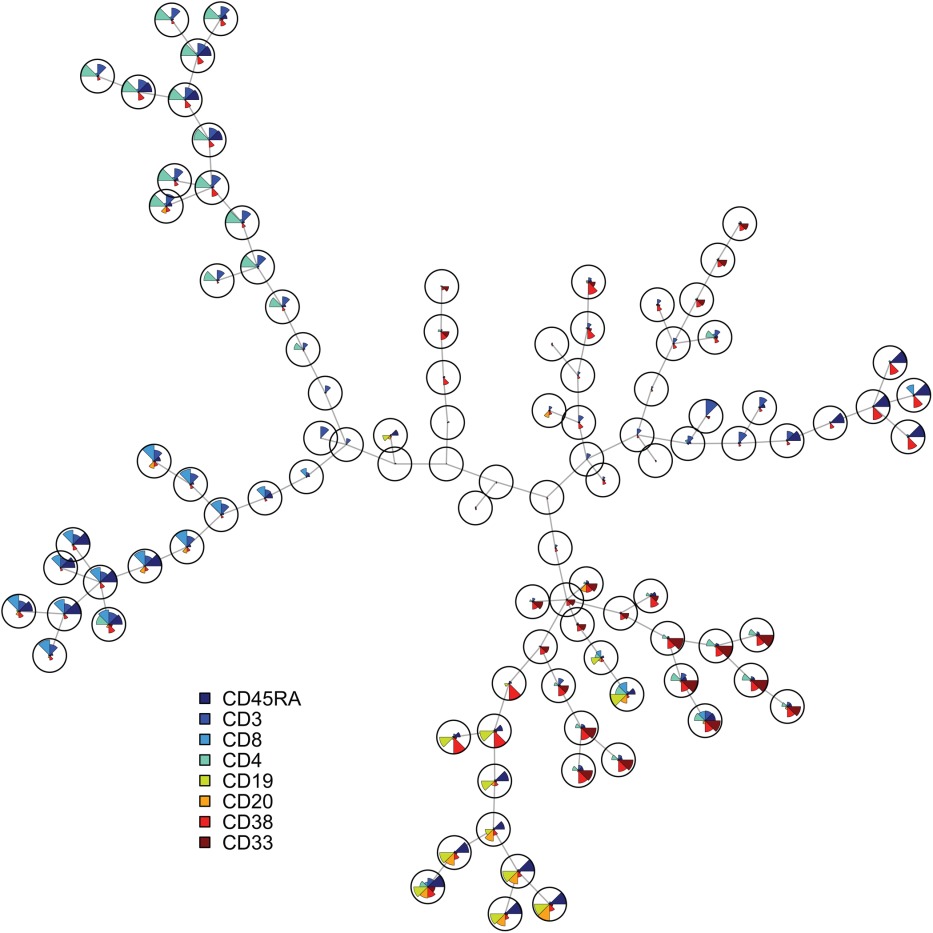
FlowSOM
Using self-organizing maps for visualization and interpretation of cytometry data
https://github.com/saeyslab/FlowSOM
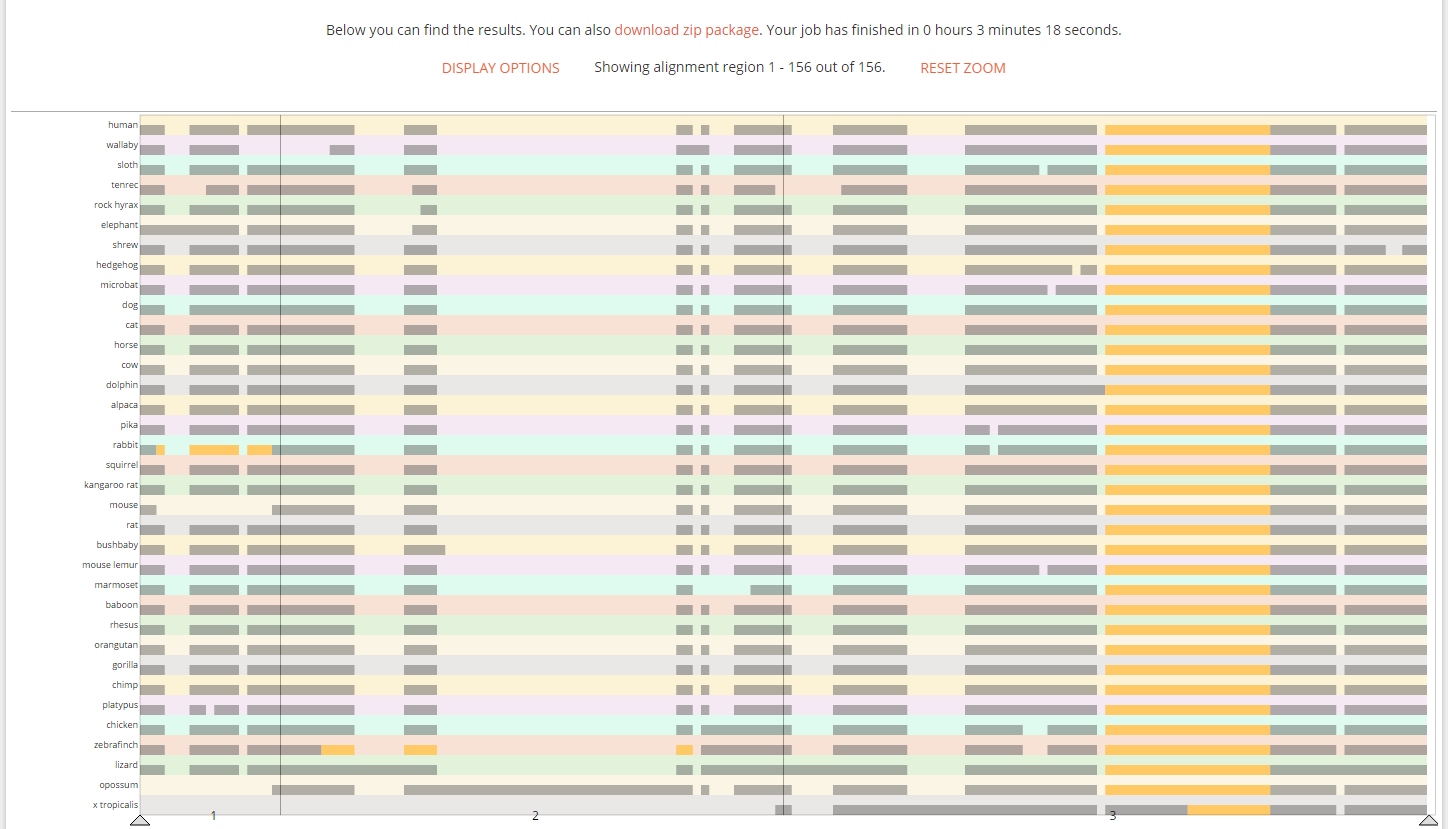
HEK293 Variant Viewer
This web tool provides for easy browsing through the sequence- and average copy-number level variations of six different HEK293 cell lines, and have included links that invoke the Integrative Genome Browser (Broad Institute) for inspection of the underlying data.
http://hek293genome.org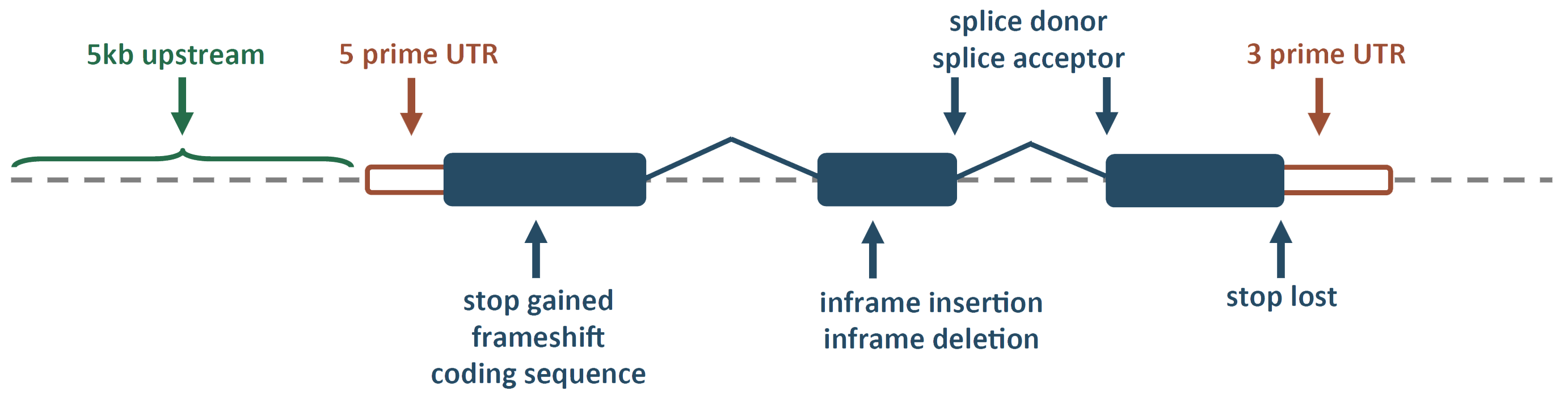
Passenger mutations finder
The Me-PuMaFind-It web tool allows scientists to easily get a list of potential passenger mutations present in any 129-derived transgenic mouse.
http://me-pamufind-it.org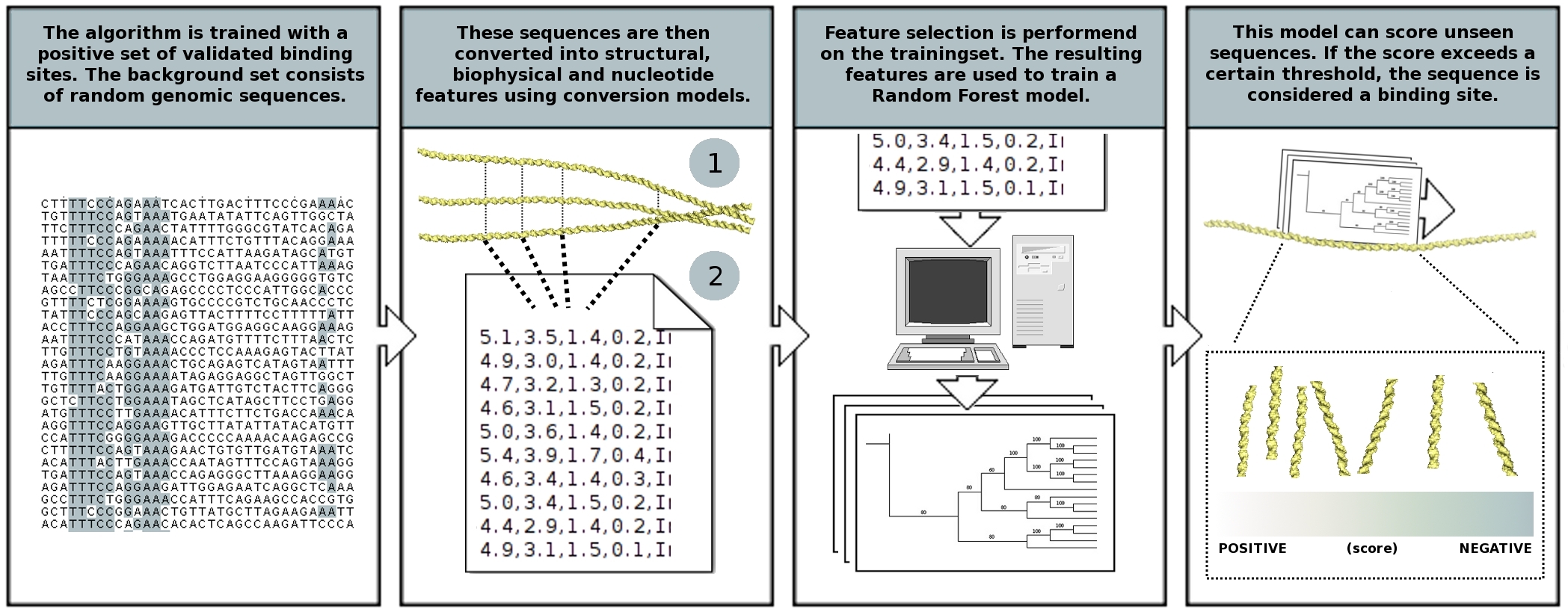
PhysBinder
A flexible integrative approach based on random forest improves prediction of transcription factor binding sites
http://bioit.irc.ugent.be/physbinder/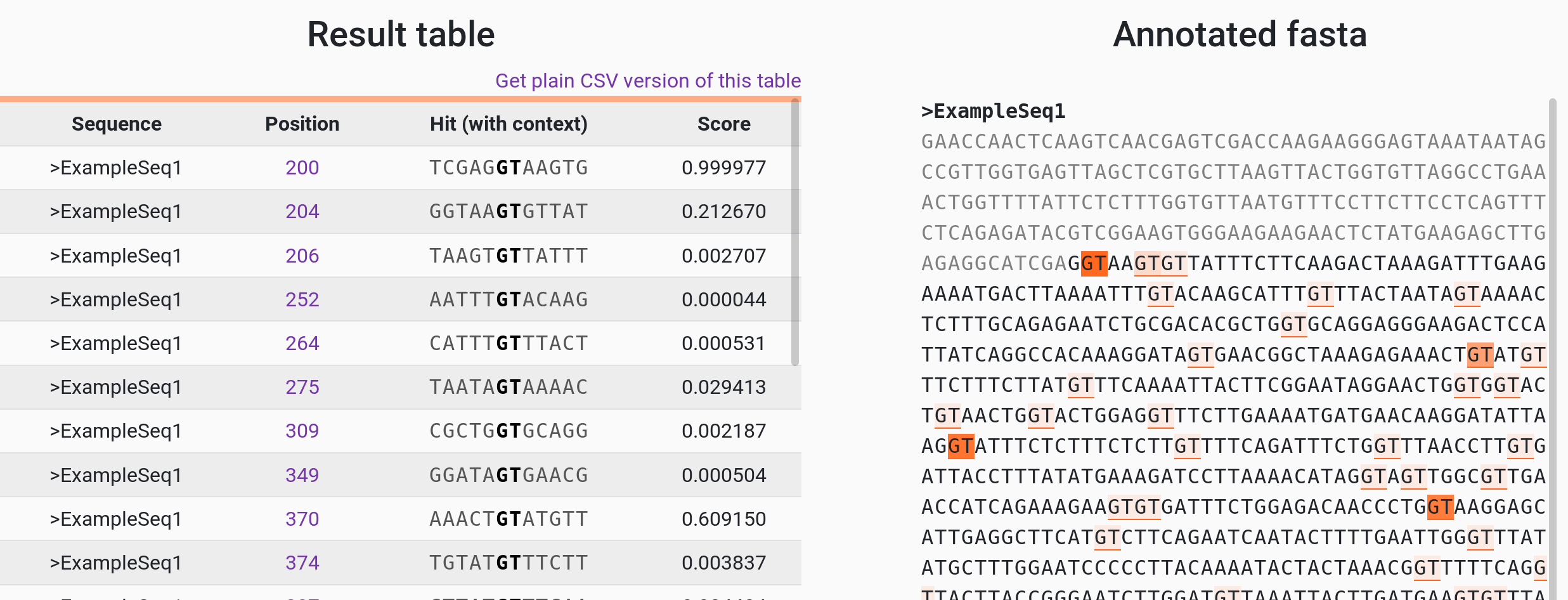
SpliceRover
SpliceRover is a prediction tool which can be used for donor and acceptor splice site prediction, in both human and arabidopsis. By making use of convolutional neural networks, we achieve state-of-the-art accuracy.
http://bioit2.irc.ugent.be/rover/splicerover/
TisRover
TISRover is a prediction tool which can be used for translation initiation site prediction in human. By making use of convolutional neural networks, we achieve state-of-the-art accuracy.
http://bioit2.irc.ugent.be/rover/tisrover/